The DeFleCT framework has been formulated and demonstrated in simulation in this paper:
Hauk O, Stenroos M. A framework for the design of flexible cross-talk functions for spatial filtering of EEG/MEG data: DeFleCT. Human Brain Mapping 2013
This page provides the
- Matlab tools that implement the DeFleCT method and enable easy visualization of data on the cortex,
- Data set that was used for making the examples in the paper
- Scripts that produce the results of the examples in the paper.
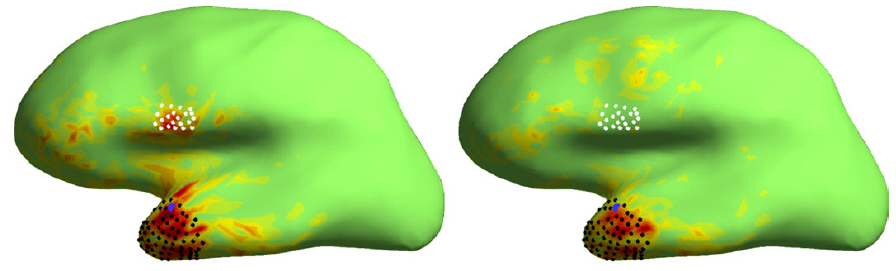
Data set and code package
For this study, we used the sample data set of MNE software that we processed according to the MNE manual: geometries for cortically-constrained source space and three-shell head model were created, sensors and the head geometry were co-registered, and a three-shell BEM model & the lead-field matrices for MEG and EEG were built. In addition, an estimate of the noise covariance matrix was constructed from pre-stimulus data. These were then imported to Matlab.
The model data can be downloaded here (24 MB).
Matlab functions and scripts that implement DeFleCT and produce the results presented in the paper can be downloaded here. Before using the codes, read conditions from readme.txt.
Further questions should be addressed to Matti Stenroos or Olaf Hauk.
For more neuroimaging-related issues, please visit our CBU Wiki pages, or have a look at recent MEG publications from the CBU.